Category: Science
Multi-Tasking Humpback Whales Sing While Feeding, Not Just Breeding

Humpback whales are famed for their songs, most often heard in breeding season when males are competing to mate with females. In recent years, however, reports of whale songs occurring outside traditional breeding grounds have become more common. A new study may help explain why.
Humpbacks sing for their supper — or at least, they sing while they hunt for it.
The research, published December 19 in PLoS ONE, uncovers the whales’ little-understood acoustic behavior while foraging.
It also reveals a previously unknown behavioral flexibility on their part that allows the endangered marine mammals to balance their need to feed continuously with the competing need to exhibit mating behaviors such as song displays.
“They need to feed. They need to breed. So essentially, they multi-task,” said study co-author Ari S. Friedlaender, research scientist at Duke University’s Nicholas School of the Environment. “This suggests the widely held behavioral dichotomy of breeding-versus-feeding for this species is too simplistic.”
Researchers from the U.S. Naval Postgraduate School, the University of California-Santa Barbara and Duke tracked 10 humpback whales in coastal waters along the Western Antarctic Peninsula in May and June 2010. The peninsula’s bays and fjords are important late-season feeding grounds where humpbacks feast on krill each austral autumn before migrating to warm-water calving grounds thousands of miles away.
Using non-invasive multi-sensor tags that attach to the whales with suction cups, the researchers recorded the whales’ underwater movements and vocalizations as they foraged.
All 10 of the tags picked up the sounds of background songs, and in two cases, they recorded intense and continuous whale singing with a level of organization and structure approaching that of a typical breeding-ground mating display. The song bouts sometimes lasted close to an hour and in one case occurred even while sensors indicated the whale, or a close companion, was diving and lunging for food.
Humpbacks sing most frequently during breeding season, but are known to sing on other occasions too, such as while escorting mother-calf pairs along migratory routes. Though the reasons they sing are still not thoroughly understood, one distinction is clear: Songs sung in breeding grounds are quite different in duration, phrase type and theme structure from those heard at other locations and times.
“The fact that we heard mating displays being sung in late-season foraging grounds off the coast of Antarctica suggests humpback whale behavior may be more closely tied to the time of year than to physical locations. This may signify an ability to engage in breeding activities outside their traditional warm-water breeding grounds,” said Douglas P. Nowacek, Repass-Rogers University Associate Professor of Conservation Technology at Duke’s Nicholas School.
As the region’s climate warms, sea ice cover around the Western Antarctic Peninsula has thinned in recent years and the water stays open later in the foraging season, he explained. Whales are remaining there longer into austral autumn to feast on krill instead of heading off to warm-water breeding grounds, as many scientists previously believed.
“Mating may now be taking place at higher latitudes,” Nowacek said. “This merits further study.”
Alison K. Stimpert, research associate in oceanography at the Naval Postgraduate School, was lead author of the new study. Lindsey E. Peavey, a PhD Student at the University of California at Santa Barbara’s Bren School of Environmental Science and Management, co-authored it with Stimpert, Friedlaender and Nowacek.
Journal Reference:
1.Stimpert AK, Peavey LE, Friedlaender AS, Nowacek DP. Humpback Whale Song and Foraging Behavior on an Antarctic Feeding Ground. PLoS One, 2012 DOI: 10.1371/journal.pone.0051214
http://www.sciencedaily.com/releases/2012/12/121219174156.htm
Scientists Construct First Detailed Map of How the Brain Organizes Everything We See

Our eyes may be our window to the world, but how do we make sense of the thousands of images that flood our retinas each day? Scientists at the University of California, Berkeley, have found that the brain is wired to put in order all the categories of objects and actions that we see. They have created the first interactive map of how the brain organizes these groupings.
The result — achieved through computational models of brain imaging data collected while the subjects watched hours of movie clips — is what researchers call “a continuous semantic space.”
“Our methods open a door that will quickly lead to a more complete and detailed understanding of how the brain is organized. Already, our online brain viewer appears to provide the most detailed look ever at the visual function and organization of a single human brain,” said Alexander Huth, a doctoral student in neuroscience at UC Berkeley and lead author of the study published Dec. 19 in the journal Neuron.
A clearer understanding of how the brain organizes visual input can help with the medical diagnosis and treatment of brain disorders. These findings may also be used to create brain-machine interfaces, particularly for facial and other image recognition systems. Among other things, they could improve a grocery store self-checkout system’s ability to recognize different kinds of merchandise.
“Our discovery suggests that brain scans could soon be used to label an image that someone is seeing, and may also help teach computers how to better recognize images,” said Huth.
It has long been thought that each category of object or action humans see — people, animals, vehicles, household appliances and movements — is represented in a separate region of the visual cortex. In this latest study, UC Berkeley researchers found that these categories are actually represented in highly organized, overlapping maps that cover as much as 20 percent of the brain, including the somatosensory and frontal cortices.
To conduct the experiment, the brain activity of five researchers was recorded via functional Magnetic Resonance Imaging (fMRI) as they each watched two hours of movie clips. The brain scans simultaneously measured blood flow in thousands of locations across the brain.
Researchers then used regularized linear regression analysis, which finds correlations in data, to build a model showing how each of the roughly 30,000 locations in the cortex responded to each of the 1,700 categories of objects and actions seen in the movie clips. Next, they used principal components analysis, a statistical method that can summarize large data sets, to find the “semantic space” that was common to all the study subjects.
The results are presented in multicolored, multidimensional maps showing the more than 1,700 visual categories and their relationships to one another. Categories that activate the same brain areas have similar colors. For example, humans are green, animals are yellow, vehicles are pink and violet and buildings are blue.
“Using the semantic space as a visualization tool, we immediately saw that categories are represented in these incredibly intricate maps that cover much more of the brain than we expected,” Huth said.
Other co-authors of the study are UC Berkeley neuroscientists Shinji Nishimoto, An T. Vu and Jack Gallant.
Journal Reference:
1.Alexander G. Huth, Shinji Nishimoto, An T. Vu, Jack L. Gallant. A Continuous Semantic Space Describes the Representation of Thousands of Object and Action Categories across the Human Brain. Neuron, 2012; 76 (6): 1210 DOI: 10.1016/j.neuron.2012.10.014
http://www.sciencedaily.com/releases/2012/12/121219142257.htm
Bullying by Childhood Peers Leaves a Trace That Can Change the Expression of a Gene Linked to Mood

A recent study by a researcher at the Centre for Studies on Human Stress (CSHS) at the Hôpital Louis-H. Lafontaine and professor at the Université de Montréal suggests that bullying by peers changes the structure surrounding a gene involved in regulating mood, making victims more vulnerable to mental health problems as they age.
The study published in the journal Psychological Medicine seeks to better understand the mechanisms that explain how difficult experiences disrupt our response to stressful situations. “Many people think that our genes are immutable; however this study suggests that environment, even the social environment, can affect their functioning. This is particularly the case for victimization experiences in childhood, which change not only our stress response but also the functioning of genes involved in mood regulation,” says Isabelle Ouellet-Morin, lead author of the study.
A previous study by Ouellet-Morin, conducted at the Institute of Psychiatry in London (UK), showed that bullied children secrete less cortisol — the stress hormone — but had more problems with social interaction and aggressive behaviour. The present study indicates that the reduction of cortisol, which occurs around the age of 12, is preceded two years earlier by a change in the structure surrounding a gene (SERT) that regulates serotonin, a neurotransmitter involved in mood regulation and depression.
To achieve these results, 28 pairs of identical twins with a mean age of 10 years were analyzed separately according to their experiences of bullying by peers: one twin had been bullied at school while the other had not. “Since they were identical twins living in the same conditions, changes in the chemical structure surrounding the gene cannot be explained by genetics or family environment. Our results suggest that victimization experiences are the source of these changes,” says Ouellet-Morin. According to the author, it would now be worthwhile to evaluate the possibility of reversing these psychological effects, in particular, through interventions at school and support for victims.
Journal Reference:
1.I. Ouellet-Morin, C. C. Y. Wong, A. Danese, C. M. Pariante, A. S. Papadopoulos, J. Mill, L. Arseneault. Increased serotonin transporter gene (SERT) DNA methylation is associated with bullying victimization and blunted cortisol response to stress in childhood: a longitudinal study of discordant monozygotic twins. Psychological Medicine, 2012; DOI: 10.1017/S0033291712002784
http://www.sciencedaily.com/releases/2012/12/121218081615.htm
Shot Away from Its Companion, Giant Star Makes Waves: Spitzer Captures Infrared Portrait

Like a ship plowing through still waters, the giant star Zeta Ophiuchi is speeding through space, making waves in the dust ahead. NASA’s Spitzer Space Telescope has captured a dramatic, infrared portrait of these glowing waves, also known as a bow shock.
Astronomers theorize that this star was once sitting pretty next to a companion star even heftier than itself. But when that star died in a fiery explosion, Zeta Ophiuchi was kicked away and sent flying. Zeta Ophiuchi, which is 20 times more massive and 80,000 times brighter than our sun, is racing along at about 54,000 mph (24 kilometers per second).
In this view, infrared light that we can’t see with our eyes has been assigned visible colors. Zeta Ophiuchi appears as the bright blue star at center. As it charges through the dust, which appears green, fierce stellar winds push the material into waves. Where the waves are the most compressed, and the warmest, they appear red. This bow shock is analogous to the ripples that precede the bow of a ship as it moves through the water, or the pileup of air ahead of a supersonic airplane that results in a sonic boom.
NASA’s Wide-field Infrared Survey Explorer, or WISE, released a similar picture of the same object in 2011. WISE sees infrared light as does Spitzer, but WISE was an all-sky survey designed to take snapshots of the entire sky. Spitzer, by contrast, observes less of the sky, but in more detail. The WISE image can be seen at: http://www.jpl.nasa.gov/news/news.php?release=2011-026 .
NASA’s Jet Propulsion Laboratory, Pasadena, Calif., manages the Spitzer Space Telescope mission for NASA’s Science Mission Directorate, Washington. Science operations are conducted at the Spitzer Science Center at the California Institute of Technology in Pasadena. Data are archived at the Infrared Science Archive housed at the Infrared Processing and Analysis Center at Caltech. Caltech manages JPL for NASA. For more information about Spitzer, visit: http://spitzer.caltech.edu and http://www.nasa.gov/spitzer .
http://www.sciencedaily.com/releases/2012/12/121218153330.htm
‘Extinct’ whale found: Odd-looking pygmy whale traced back 2 million years

The pygmy right whale, a mysterious and elusive creature that rarely comes to shore, is the last living relative of an ancient group of whales long believed to be extinct, a new study suggests.
The findings, published Tuesday, Dec. 18, in the Proceedings of the Royal Society B, may help to explain why the enigmatic marine mammals look so different from any other living whale.
“The living pygmy right whale is, if you like, a remnant, almost like a living fossil,” said Felix Marx, a paleontologist at the University of Otago in New Zealand. “It’s the last survivor of quite an ancient lineage that until now no one thought was around.”
The relatively diminutive pygmy right whale, which grows to just 21 feet (6.5 meters) long, lives out in the open ocean. The elusive marine mammals inhabit the Southern Hemisphere and have only been spotted at sea a few dozen times. As a result, scientists know almost nothing about the species’ habits or social structure.
The strange creature’s arched, frownlike snout makes it look oddly different from other living whales. DNA analysis suggested pygmy right whales diverged from modern baleen whales such as the blue whale and the humpback whale between 17 million and 25 million years ago. However, the pygmy whales’ snouts suggested they were more closely related to the family of whales that includes the bowhead whale. Yet there were no studies of fossils showing how the pygmy whale had evolved, Marx said.
To understand how the pygmy whale fit into the lineage of whales, Marx and his colleagues carefully analyzed the skull bones and other fossil fragments from pygmy right whales and several other ancient cetaceans.
The pygmy whale’s skull most closely resembled that of an ancient family of whales called cetotheres that were thought to have gone extinct around 2 million years ago, the researchers found. Cetotheres emerged about 15 million years ago and once occupied oceans across the globe.
The findings help explain how pygmy whales evolved and may also help shed light on how these ancient “lost” whales lived. The new information is also a first step in reconstructing the ancient lineage all the way back to the point when all members of this group first diverged, he said.
Thanks to Dr. Lutter for bringing this to the attention of the It’s Interesting community.
Do Palm Trees Hold the Key to Immortality?
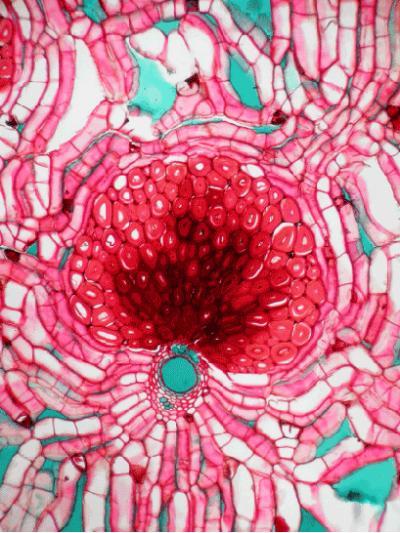
For centuries, humans have been exploring, researching, and, in some cases, discovering how to stave off life-threatening diseases, increase life spans, and obtain immortality. Biologists, doctors, spiritual gurus, and even explorers have pursued these quests — one of the most well-known examples being the legendary search by Ponce de León for the “Fountain of Youth.” Yet the key to longevity may not lie in a miraculous essence of water, but rather in the structure and function of cells within a plant — and not a special, mysterious, rare plant, but one that we may think of as being quite commonplace, even ordinary: the palm.
As an honors botany student at the University of Leeds, P. Barry Tomlinson wrote a prize-winning essay during his final year titled, “The Span of Life.” Fifty years later, Tomlinson (now a Distinguished Professor at The Kampong Garden of the National Tropical Botanical Garden, Miami, FL) teamed up with graduate student Brett Huggett (Harvard University, MA) to write a review paper exploring the idea that palms may be the longest-lived tree, and whether this might be due to genetic underpinnings. Having retained his essay in his personal files, Tomlinson found that it provided an excellent literature background for working on the question of cell longevity in relation to palms. Together, Tomlinson and Huggett published their review in the December issue of the American Journal of Botany.
A component of an organism’s life span that biologists have been particularly interested in is whether longevity is genetically determined and adaptive. For botanists, discovering genetic links to increasing crop production and the reproductive lifespan of plants, especially long-lived ones such as trees, would be invaluable.
In their paper, Tomlinson and Huggett emphasize that in many respects, an organisms’ life span, or longevity, is determined by the period of time in which its cells remain functionally metabolically active. In this respect, plants and animals differ drastically, and it has to do with how they are organized — plants are able to continually develop new organs and tissues, whereas animals have a fixed body plan and are not able to regenerate senescing organs. Thus, plants can potentially live longer than animals.
“The difference in potential cell longevity in plants versus animals is a significant point,” states Tomlinson. “It is important to recognize that plants, which are so often neglected in modern biological research, can be informative of basic cell biological features in a way that impacts human concern at a fundamental level.”
The authors focused their review on palm trees because palms have living cells that may be sustained throughout an individual palm’s lifetime, and thus, they argue, may have some of the longest living cells in an organism. As a comparison, in most long-lived trees, or lignophytes, the main part, or trunk, of the tree is almost entirely composed of dead, woody, xylem tissues, and in a sense is essentially a supportive skeleton of the tree with only an inner ring of actively dividing cells. For example, the skeleton of Pinus longaeva may be up to 3000 years old, but the active living tissues can only live less than a century.
In contrast, the trunks of palms consist of cells that individually live for a long time, indeed for the entire life of an individual.
Which brings up the question of just how long can a palm tree live? The authors point out that palm age is difficult to determine, primarily because palms do not have secondary growth and therefore do not put down annual or seasonal growth rings that can easily be measured. However, age can be quite accurately assessed based on rate of leaf production and/or visible scars on the trunk from fallen leaves. Accordingly, the authors found that several species of palm have been estimated to live as long as 100 and even up to 740 years. The important connection here is that while the “skeleton” of the palm may not be as old as a pine, the individual cells in its trunk lived, or were metabolically active, as long as, or longer than those of the pine’s.
Most plants, in addition to increasing in height as they age, also increase in girth, putting down secondary vascular tissue in layers both on the inner and outer sides of the cambium as they grow. However, palms do not have secondary growth, and there is no addition of secondary vascular tissue. Instead, stem tissues are laid down in a series of interconnected vascular bundles — thus, not only is the base of the palm the oldest and the top the youngest, but these tissues from old to young, from base to top, must also remain active in order to provide support and transport water and nutrients throughout the tree.
Indeed, the authors illustrate this by reviewing evidence of sustained primary growth in two types of palms, the coconut and the sago palm. These species represent the spectrum in tissue organization from one where cells are relatively uniform and provide both hydraulic and mechanical functions (the coconut) to one where these functions are sharply divided with the inner cells functioning mainly for transporting water and nutrients and the outer ones for mechanical support (the sago palm). This represents a progression in specialization of the vascular tissues.
Moreover, there is evidence of continued metabolic activity in several types of tissues present in the stems of palms, including vascular tissue, fibers, ground tissue, and starch storage. Since the vascular tissues in palms are nonrenewable, they must function indefinitely, and Tomlinson and Huggett point out that sieve tubes and their companion cells are remarkable examples of cell longevity as they maintain a long-distance transport function without replacement throughout the life of the stem, which could be for centuries.
Despite several unique characteristics of palms, including the ability to sustain metabolically active cells in the absence of secondary tissues, seemingly indefinitely, unlike conventional trees, in which metabolically active cells are relatively short-lived, the authors do not conclude that the extended life span of palms is genetically determined.
“We are not saying that palms have the secret of eternal youth, and indeed claim no special chemical features which allows cells in certain organisms to retain fully differentiated cells with an indefinite lifespan,” states Tomlinson. “Rather, we emphasize the distinctive developmental features of palm stems compared with those in conventional trees.”
Tomlinson indicates that this reflects the neglect of the teaching of palm structure in modern biology courses. “This paper raises incompletely understood aspects of the structure and development of palms, emphasizing great diversity in these features,” he concludes. “This approach needs elaborating in much greater detail, difficult though the subject is in terms of conventional approaches to plant anatomy.”
Journal Reference:
1.P. B. Tomlinson, B. A. Huggett. Cell longevity and sustained primary growth in palm stems. American Journal of Botany, 2012; 99 (12): 1891 DOI: 10.3732/ajb.1200089
http://www.sciencedaily.com/releases/2012/12/121219092842.htm
Hatching Order Influences Birds’ Behavior

The hatching order of birds influences how they behave in adult life according to research from the Lancaster Environment Centre. Dr Ian Hartley and Dr Mark Mainwaring (LEC) are the authors of the study in Animal Behaviour, which looked at how the birds’ behaviour was affected by the way their parents cared for them as hatchlings.
They found that the youngest members of zebra finch broods are more adventurous than their older siblings in adult life.
Dr Hartley said that the study showed for the first time that hatching order influences birds’ “behavioural repertoires” in adulthood.
Hatching eggs over a period of time, rather than all at once, is known as “hatching asynchrony” and occurs when eggs are incubated as soon as they are laid. For a zebra finch, this means that birds born up to four days apart can share the same nest and must compete for food.
The researchers experimentally controlled hatching synchrony within clutches, so that some clutches hatched simultaneously, while others hatched over a period of days. They then tested the behaviour of over one hundred offspring as adults. They found the youngest birds from asynchronously hatched clutches explored their environment more widely.
They measured how explorative the zebra finches were by recording how many times they visited bird feeders within an unfamiliar test aviary. They found that the youngest offspring in a brood approached the feeders significantly more often than their peers within a 30 minute period.
Researchers wanted to know how the method of rearing affected the behaviour of offspring beyond the nest, once they were living as independent adult birds. The results have implications for understanding how environmental stability might influence behaviours, and how flexible animals might be at coping with environmental change.
Journal Reference:
1.Mark C. Mainwaring, Ian R. Hartley. Hatching asynchrony and offspring sex influence the subsequent exploratory behaviour of zebra finches. Animal Behaviour, 2012; DOI: 10.1016/j.anbehav.2012.10.009
http://www.sciencedaily.com/releases/2012/12/121207094343.htm
The brain’s natural valium

Hitting the wall in the middle of a busy work day is nothing unusual, and a caffeine jolt is all it takes to snap most of us back into action. But people with certain sleep disorders battle a powerful urge to doze throughout the day, even after sleeping 10 hours or more at night. For them, caffeine doesn’t touch the problem, and more potent prescription stimulants aren’t much better. Now, a study with a small group of patients suggests that their condition may have a surprising source: a naturally occurring compound that works on the brain much like the key ingredients in chill pills such as Valium and Xanax.
The condition is known as primary hypersomnia, and it differs from the better known sleep disorder narcolepsy in that patients tend to have more persistent daytime sleepiness instead of sudden “sleep attacks.” The unknown cause and lack of treatment for primary hypersomnia has long frustrated David Rye, a neurologist at Emory University in Atlanta. “A third of our patients are on disability,” he says, “and these are 20- and 30-year-old people.”
Rye and colleagues began the new study with a hunch about what was going on. Several drugs used to treat insomnia promote sleep by targeting receptors for GABA, a neurotransmitter that dampens neural activity. Rye hypothesized that his hypersomnia patients might have some unknown compound in their brains that does something similar, enhancing the activity of so-called GABAA receptors. To try to find this mystery compound, he and his colleagues performed spinal taps on 32 hypersomnia patients and collected cerebrospinal fluid (CSF), the liquid that bathes and insulates the brain and spinal cord. Then they added the patients’ CSF to cells genetically engineered to produce GABAA receptors, and looked for tiny electric currents that would indicate that the receptors had been activated.
In that first pass, nothing happened. However, when the researchers added the CSF and a bit of GABA to the cells, they saw an electrical response that was nearly twice as big as that caused by GABA alone. All of this suggests that the patients’ CSF doesn’t activate GABAA receptors directly, but it does make the receptors almost twice as sensitive to GABA, the researchers report today in Science Translational Medicine. This effect is similar to that of drugs called benzodiazepines, the active ingredients in antianxiety drugs such as Valium. It did not occur when the researchers treated the cells with CSF from people with normal sleep patterns.
Follow-up experiments suggested that the soporific compound in the patients’ CSF is a peptide or small protein, presumably made by the brain, but otherwise its identity remains a mystery.
The idea that endogenous benzodiazepinelike compounds could cause hypersomnia was proposed in the early 1990s by Elio Lugaresi, a pioneering Italian sleep clinician, says Clifford Saper, a neuroscientist at Harvard Medical School in Boston. But several of Lugaresi’s patients later turned out to be taking benzodiazepines, which undermined his argument, and the idea fell out of favor. Saper says the new work makes a “pretty strong case.”
Based on these results, Rye and his colleagues designed a pilot study with seven patients using a drug called flumazenil, which counteracts benzodiazepines and is often used to treat people who overdose on those drugs. After an injection of flumazenil, the patients improved to near-normal levels on several measures of alertness and vigilance, the researchers report. Rye says these effects lasted up to a couple hours.
In hopes of longer-lasting benefits, the researchers persuaded the pharmaceutical company Hoffmann-La Roche, which makes the drug, to donate a powdered form that can be incorporated into dissolvable tablets taken under the tongue and a cream applied to the skin. One 30-something patient has been taking these formulations for 4 years and has improved dramatically, the researchers report in the paper. She has resumed her career as an attorney, from which her hypersomnia had forced her to take a leave of absence.
The findings are “certainly provocative,” Saper says, although they’ll have to be replicated in a larger, double-blind trial to be truly convincing.
Even so, says Phyllis Zee, a neurologist at Northwestern University in Evanston, Illinois: “This gives us a new window into thinking about treatments” for primary hypersomnia. “These patients don’t respond well to stimulants,” Zee says, so a better strategy may be to inhibit the sleep-promoting effects of GABA—or as Rye puts it, releasing the parking brake instead of pressing the accelerator.
The next steps are clear, Rye says: Identify the mystery compound, figure out a faster way to detect it, and conduct a larger clinical trial to test the benefits of flumazenil. However, the researchers first need someone to fund such a study. So far, Rye says, they’ve gotten no takers.
http://news.sciencemag.org/sciencenow/2012/11/putting-themselves-to-sleep.html
