
When most people hear about electroconvulsive therapy, or ECT, it typically conjures terrifying images of cruel, outdated and pseudo-medical procedures. Formerly known as electroshock therapy, this perception of ECT as dangerous and ineffective has been reinforced in pop culture for decades – think the 1962 novel-turned-Oscar-winning film “One Flew Over the Cuckoo’s Nest,” where an unruly patient is subjected to ECT as punishment by a tyrannical nurse.
Despite this stigma, ECT is a highly effective treatment for depression – up to 80% of patients experience at least a 50% reduction in symptom severity. For one of the most disabling illnesses around the world, I think it’s surprising that ECT is rarely used to treat depression.
Contributing to the stigma around ECT, psychiatrists still don’t know exactly how it heals a depressed person’s brain. ECT involves using highly controlled doses of electricity to induce a brief seizure under anesthesia. Often, the best description you’ll hear from a physician on why that brief seizure can alleviate depression symptoms is that ECT “resets” the brain – an answer that can be fuzzy and unsettling to some.
As a data-obsessed neuroscientist, I was also dissatisfied with this explanation. In our newly published research, my colleagues and I in the lab of Bradley Voytek at UC San Diego discovered that ECT might work by resetting the brain’s electrical background noise.
Listening to brain waves
To study how ECT treats depression, my team and I used a device called an electroencephalogram, or EEG. It measures the brain’s electrical activity – or brain waves – via electrodes placed on the scalp. You can think of brain waves as music played by an orchestra. Orchestral music is the sum of many instruments together, much like EEG readings are the sum of the electrical activity of millions of brain cells.
Two types of electrical activity make up brain waves. The first, oscillations, are like the highly synchronized, melodic music you might hear in a symphony. The second, aperiodic activity, is more like the asynchronous noise you hear as musicians tune their instruments. These two types of activities coexist in the brain, together creating the electrical waves an EEG records.
Importantly, tuning noises and symphonic music shouldn’t be mistaken for one another. They clearly come from different processes and serve different purposes. The brain is similar in this way – aperiodic activity and oscillations are different because the biology driving them is distinct.
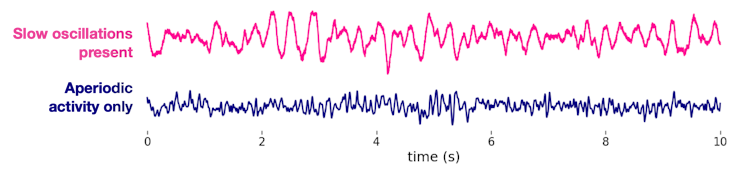
However, the methods neuroscientists have traditionally used to analyze these signals are unable to differentiate between the oscillations (symphony) and the aperiodic activity (tuning). Both are critical for the orchestra, but so far neuroscientists have mostly ignored – or entirely missed – aperiodic signals because they were thought to be just the brain’s background noise.
In our new research, my team and I show that ignoring aperiodic brain activity likely explains the confusion behind about how ECT treats depression. It turns out we’ve been missing this signal all along.
Connecting aperiodic activity and ECT
Since the 1940s, ECT has been associated with increases in slow oscillations in the brain waves of patients. However, those slow oscillations have never been linked to how ECT works. The degree to which slow oscillations appear is not consistently related to how much symptoms improve following ECT. Nor have ideas about how the brain produces slow oscillations connected those processes to the pathology underlying depression.
Because these two types of brain waves are difficult to separate in measurements, I wondered if these slow oscillations were in fact incorrectly measured aperiodic activity. Returning to our orchestra analogy, I believed that scientists had misidentified the tuning sounds as symphony music.
To investigate this, my team and I gathered three EEG datasets: one from nine patients with depression undergoing ECT in San Diego, another from 22 patients in Toronto receiving ECT and a third from 22 patients in Toronto participating in a clinical trial of magnetic seizure therapy, or MST, a newer alternative to ECT that starts a seizure with magnets instead of electricity.
We found that aperiodic activity increases by more than 40% on average following ECT. In patients who received MST treatment, aperiodic activity increases more modestly, by about 16%. After accounting for changes in aperiodic activity, we found that slow oscillations do not change much at all. In fact, slow oscillations were not even detected in some patients, and aperiodic activity dominated their EEG recordings instead.
How ECT treats depression
But what does aperiodic activity have to do with depression?
A long-standing theory of depression states that severely depressed patients have too few of a type of brain cell called inhibitory cells. These cells can turn other brain cells on and off, and maintaining the balance of these on and off states is critical for healthy brain function. This balance is particularly relevant for depression because the brain’s ability to turn cells off plays an important role in how it responds to stress, a function that, when not working properly, makes people particularly vulnerable to depression.
Using a mathematical model of cell type-based electrical activity, I linked increases in aperiodic activity, like those seen in the ECT patients, to a huge change in the activity of these inhibitory cells. This change in aperiodic activity may be restoring the crucial on and off balance in the brain to a healthy level.
Even though scientists have been recording EEGs from ECT patients for decades, this is the first time that brain waves have been connected to this particular brain malfunction.
Altogether, though our sample size is relatively small, our findings indicate that ECT and MST likely treat depression by resetting aperiodic activity and restoring the function of inhibitory brain cells. Further study can help destigmatize ECT and highlight new directions for the research and development of depression treatments. Listening to the nonmusical background noise of the brain could help solve other mysteries, like how the brain changes in aging and in illnesses like schizophrenia and epilepsy.











